Optimizing GB Models for Medical Diagnosis
About the project
![]()
![]()
This project compared the performance of three gradient boosting algorithms (XGBoost, LightGBM, and CatBoost) on a binary classification task for medical diagnosis. The main focus was on improving prediction performance through hyperparameter and threshold optimization.
Link to the project files: github.com/Al-1n/Gradient_Boosting_Tuning
Full scientific report: GBT Classification Report
Requirements
| Optuna | Hyperopt | LightGBM | CatBoost |
|---|---|---|---|
| XGBoost | IpyWidgets | Imblearn | Networkx |
| CloudPickle | Plotly | SQLAlchemy | ImageIO |
| PyWavelets | TiffFile | Pandas | Numpy |
| Seaborn | Matplotlib | Math | Sklearn |
| PyGraphViz | IPython | JupyterLab |
How to use this project
The code in these files can be adapted to perform similar tests and comparisons for different datasets and models.
Any environment that can load a python kernel and run jupyter notebooks such as vs code, google collab or conda can be used.
Contributors
Data and Challenges
The analysis utilizes a popular sample of the Pima Indians Diabetes Database, which presents several challenges:
- Small sample size
- Numerous missing measurements
- Class imbalance
To address these, the project employs model-based imputations, synthetic sampling for the minority class, and feature selection to refine the dataset.
Methods
1. Data Preprocessing:
- Model-based imputations using LightGBM
- Feature scaling
- ADASYN for data augmentation
- Feature selection with SelectKBest
- Class weighting
2. Model Training and Optimization:
- Baseline models without optimization
- Hyperparameter optimization using Optuna and Hyperopt
- Decision threshold optimization with TunedThresholdClassifierCV
Performance Analysis
Recall Performance
In medical diagnosis, recall (true positive rate) is crucial. Recall scores ranged from 0.64 to 0.89, with significant improvements observed through optimization.
- XGBoost: Highest recall achieved with Opt+Th optimization.
- LightGBM: Similar improvement pattern, with Opt+Th providing the best recall.
- CatBoost: Strong recall even at baseline, with Opt+Th yielding the highest recall.
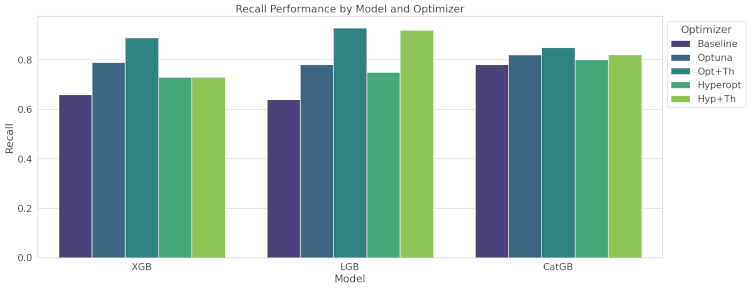
The grouped bar graph shows the recall performance of the three gradient boosting models (XGBoost, LightGBM, and CatBoost) across different optimization methods.
F1-Score Performance
The F1-score balances precision and recall, critical for imbalanced datasets.
- XGBoost: Opt+Th optimization significantly improved the F1-score.
- LightGBM: Similar improvement with Opt+Th optimization yielding the highest F1-score.
- CatBoost: Strong baseline performance, with highest F1-score achieved through Opt+Th.
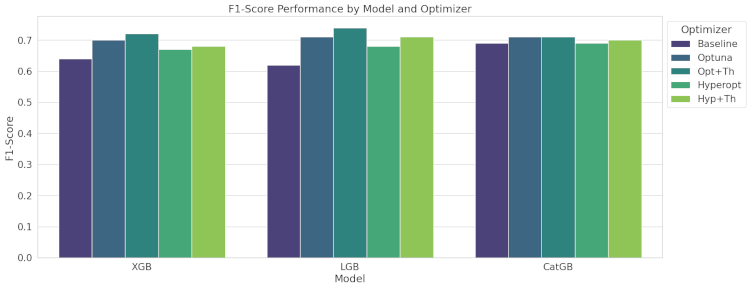
The grouped bar graph shows the F1-score performance of the three gradient boosting models (XGBoost, LightGBM, and CatBoost) across different optimization methods.
AUROC and AUPRC
AUROC (Area Under the Receiver Operating Characteristic Curve):
- XGBoost and LightGBM: High AUROC scores (0.78 to 0.83), with Optuna, Opt+Th, and Hyp+Th optimizers achieving the best results.
- CatBoost: Highest AUROC scores, maintaining around 0.83 with optimizations.
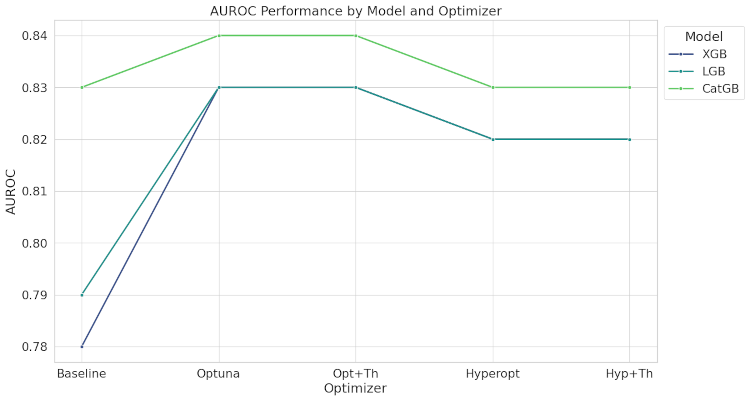
The line plot shows the AUROC variation across different optimization methods for the three gradient boosting models.
AUPRC (Area Under the Precision-Recall Curve):
- XGBoost: Scores improved to 0.62 with Optuna and Opt+Th optimizers.
- LightGBM: Highest scores achieved with Optuna and Opt+Th (0.62).
- CatBoost: Leading with an AUPRC of 0.65, even at baseline.
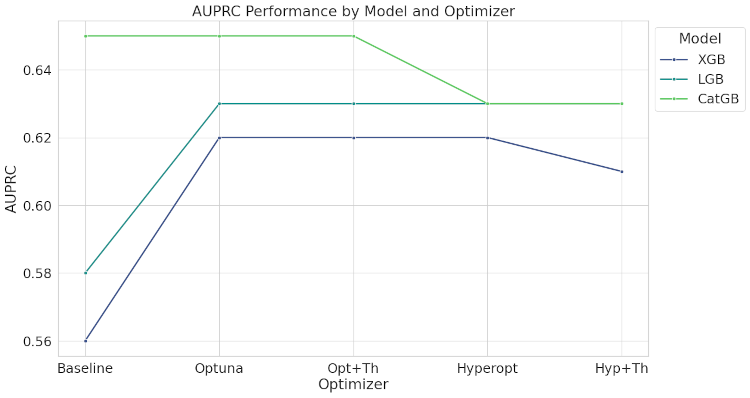
The line plot shows the AUPRC variation across different optimization methods for the three gradient boosting models.
Conclusions
- Optimization techniques generally improved model performance across all metrics.
- CatBoost demonstrated strong performance even with baseline parameters.
- XGBoost and LightGBM benefited most from optimization, showing the largest improvements.
- For scenarios prioritizing recall, such as medical diagnosis, optimized XGBoost and LightGBM models performed best.
- CatBoost consistently outperformed in AUROC and AUPRC, indicating superior overall performance.
Future Work
Further testing is recommended to confirm these findings and explore their applicability to other datasets and domains.